Artificial intelligence and machine learning is revolutionizing protein design, which opens many new applications in neuroscience that we have only dreamt of thus far ranging from therapy to basic research. We seek to exploit these technical developments to develop proteins that bind proteins that are important in memory and learning which allow us to manipulate their function.
Protein de novo design requires access to high-through put biophysical experiments for experimental characterization. We are very well positioned for these experiments, as Magnus is the coordinator of the “Biophysics and Biochemistry core facility” at the Department of Molecular Biology and Genetics.
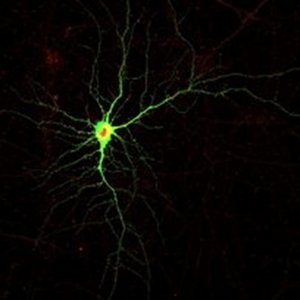
How are memories stored in out brains? This is the central question in the Center of Excellence PROMEMO of which we are part. Decades of research in molecular neuroscience has shown that activity-dependent changes in synaptic strengths and connections are the cellular and molecular underpinning. This strengthening requires that the molecular machinery of neurotransmission is recruited to specific synapses in response to the biochemical activity triggered by synaptic stimulation. But how do these mechanisms work at the molecular level? Unsurprisingly, intrinsically disordered proteins seem to play a key role. We aim to answer this question by studying:
Disordered tails of neuronal receptors: Many crucial neuronal receptors have long disordered tails that are not part of their main receptor function, but rather regulate their location, interactions and signaling output. We study such receptor tails in vitro and in cells.
Reverse engineering activity dependent protein recruitment: Imagine if we could target any protein to a synapse in response to synaptic stimulation. This is what we are trying to achieve by reverse engineering the mechanisms neurons use to target proteins to the right synapses.
Cells are divided into compartment with different functions. In addition to the traditional membrane-bounded organelles, it has recently been discovered that there are another class of “membrane-less organelles” that arise due to reversible self-assembly of macromolecules. Intrinsically disordered proteins play a key role in formation of such biomolecular condensates. We study the effects of biomolecular condensates particularly on enzymatic reactions, which can be dramatically increase or decreased when transferred into a condensate. Currently, we focus on two different types of enzymatic systems:
Signaling pathways: Kinases and phosphatases are the backbone of cellular signaling. Many signaling enzymes are found in biomolecular condensates, but we know little of how the environment of the condensate affects their activities. Understanding such effects are crucial to the emergent class of pharmaceuticals that aim to modulate biomolecular condensates.
Biosynthetic pathways: Nature has created a treasure trove of organic compounds with potential applications in medicine and industry. Many of these compounds are too rare to extract from natural sources and has to be created in a lab – typically in genetically engineered microorganisms. However, many biosynthetic pathways work poorly when transplanted to e.g. yeast or bacteria. We aim to develop biomolecular condensates as “designer organelles” for biosynthesis pathways of complex natural compounds.